Introduction
Nosema is a pathogen of concern for beekeepers because it has been shown to adversely affect honey bee health. There are two species of Nosema known to impact the health of honey bees, Nosema apis and Nosema ceranae. Currently, fewer Nosema apis infections are found Nosema ceranae has become the dominant microsporidian infection in western honey bee colonies (Fries 2010). Microsporidia are single-celled organisms that parasitize animal hosts via the dispersal of spores. Nosema ceranae spores are slightly smaller than spores of Nosema apis. However, the two species are nearly impossible to differentiate without molecular tools. Therefore, we generically refer to infections of either or both species as Nosema infections.
Nosema is a microsporidian that represents a taxonomic group of single-celled fungal parasites known to infect various animal hosts. Honey bees contract the disease by ingesting Nosema spores through trophallaxis (food sharing) or by ingesting fecal waste from an infected nest mate. Once ingested, the Nosema replicates inside of midgut (stomach) cells and hijacks nutrition from the honey bee. Eventually the Nosema spores replicate so much that they rupture the midgut cell membrane. This releases more spores into the honey bee's midgut and increases the likelihood that the disease will be transmitted to other bees.
Infected honey bees have shortened lifespans and commonly demonstrate pronounced hunger (i.e. begging to be fed by other adult honey bees more often; Mayak and Naug 2009). The risk of Nosema infection can be particularly unsettling to beekeepers because colonies often do not show outward manifestations of infection until the colony is severely diminished.
Diagnosis of Nosema infection is traditionally performed using light microscopy, and the infection level is estimated using a hemocytometer to count spores. The materials needed and steps to quantify Nosema infection in a honey bee colony are described below.
Field Collections
Necessary Supplies
- Screw-cap containers (e.g., half-pint mason jar, baby-food jar, or any other container that holds approximately 8 fluid ounces or 250 ml)—one per colony to be sampled
- Isopropyl alcohol (i.e., rubbing alcohol)
- Disposable baking pan or plastic tub (approximately 22 × 33 cm or 9 × 11 inch)
Overview
You will need to collect a sample of approximately 100 adult worker bees (about ¼ cup of bees) from each colony from which you intend to quantify the Nosema infestation. It is best to collect adult bees that represent a mix of adult bee ages to ensure that your sampling is not biased by the age of the workers. This can best be achieved by collecting workers from a frame with open brood present.
It will help you stay organized if each colony has a unique identification number or name, especially if you are collecting numerous samples at once. You may also find a notebook useful as you collect samples to record information such as the date, apiary name, colony ID, and/or colony condition.
Detailed Instructions
- Label your collection container with the date, and colony ID (possibly the apiary name if you maintain multiple apiaries).
- Fill the collection container approximately two thirds full with isopropyl alcohol.
- Collect about 100 adult bees.
- Shake the bees from a brood frame containing open brood into a disposable plastic bin or baking pan. *Note: if you happen to pull a frame and see the queen on it, put it back gently and select another frame.
- Gently shake the pan to collect all of the bees into one corner.
- Pour the bees into the prepared collection container.
4. Seal the collection container and transport it to an area equipped for Nosema quantification (see the "Nosema Quantification" section, next).
5. Repeat steps 1–4 for each colony.
Nosema Quantification
Necessary Supplies
This procedure requires scientific equipment that is not likely to be found at a local retailer. Fisher Scientific (http://www.fishersci.com) is an online supplier that carries an array of scientific equipment including microscopes, hemocytometers, gloves, graduated cylinders, etc. You may find this supplier or other web-based scientific suppliers helpful in acquiring all of the supplies necessary to quantify Nosema spores. Furthermore, some of the required supplies (e.g., a compound microscope) may be cost prohibitive for an individual beekeeper. However, Nosema quantification can be more affordable if several beekeepers or beekeeping clubs purchase these larger items to share.
- Latex or nitrile gloves
- Paper towels
- 1 quart (~1000 ml) zip-locking bag (one for each colony sampled)
- Rolling pin
- Graduated cylinder, or pipette (something to measure 100 ml)
- Water
- Improved neubauer hemocytometer
- Glass cover slips (one for each colony sampled)
- Single-use dropper or pipettor w/ disposable pipette tips
- Compound microscope with up to 400× magnification.
- No-lint tissue to clean microscope and hemocytometer surfaces (e.g., kimwipes)
- Isopropyl alcohol (i.e., rubbing alcohol)
Overview
To quantify the Nosema infection rate (number of Nosema spores per bee) for a colony, you will crush a known number of forager bees in 1 ml of water per bee. In the description below, we recommend crushing 100 bees in 100 ml of water. Next, place a small sample of the liquid produced by crushing the bees in the water onto a hemocytometer and visualize it under a compound microscope. You will then count the total number of spores that you see in 5 of the grid blocks and use that number (your raw spore count) to calculate the number of spores/bee. *Note: we recommend wearing latex or nitrile gloves while handling the dead bees soaked in alcohol.
- While wearing gloves, count 100 intact worker bees from a single colony sample.
- Place the bees onto a paper towel to dry excess liquid from the bees.
- Transfer the 100 bees into a zip-locking bag and seal the bag (ensure that as much air as possible is removed from the bag).
- Crush the bees within the zip-locking bag by placing the bag onto a hard flat surface and carefully rolling with a rolling pin. Take care to crush all of the bees in the bag without tearing any holes in the bag (Figure 1).
- Add 100 ml of water to the bag of crushed bees. *Note: You should always add 1 mL of water per bee, so if you started with 80 bees instead of 100 you will need to adjust the amount of water that you add to 80ml instead of 100ml.
- Shake the bag for approximately 30 seconds to mix well (Figure 1).
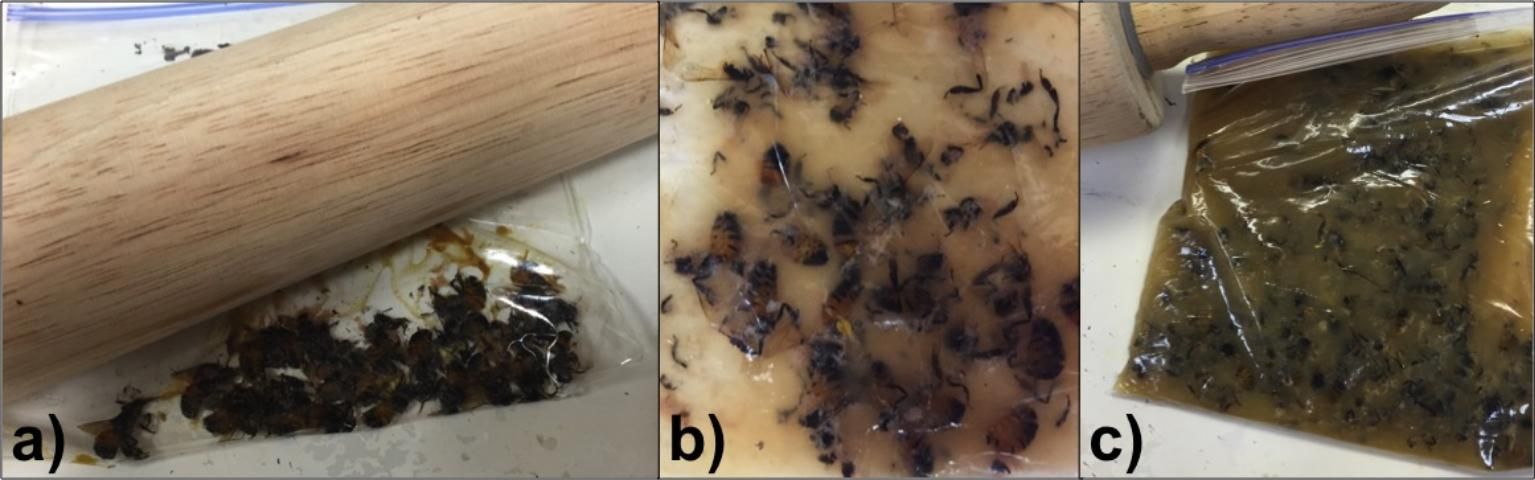
Credit: Liana Teigen (honeybee@ifas.ufl.edu), University of Florida.
7. Place a glass coverslip over the grid of the hemocytometer.
8. Use the disposable dropper or pipettor with disposable pipette tips to place a drop of the sample into the well of the hemocytometer (just at the edge of the cover slip; Figure 2).

Credit: UF/IFAS Honey Bee Lab
9. Allow the sample to settle on the hemocytometer for 2–3 minutes.
10. Place the hemocytometer on a microscope and zoom to 400x (Figure 3).
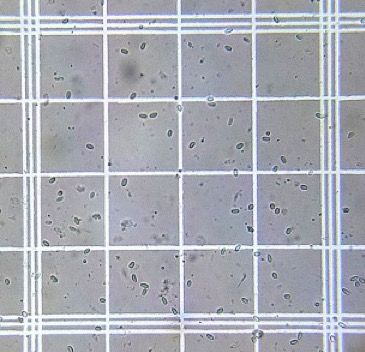
Credit: Used with permission from Randy Oliver (http://scientificbeekeeping.com)
11. Count the total number of spores present in five blocks on the hemocytometer grid (Figure 4).
- There are 25 primary blocks that each contain 16 smaller squares on the hemocytometer grid. Nine of those blocks have been numbered in Figure 4.
- Count the total number of the spores present in blocks 1–5.
- If one of the blocks happens to be obscured by debris (e.g., block number 3) you can skip that block and count the spores in block 6 instead. Blocks 7–9 can be used as well if multiple blocks are obscured from view.
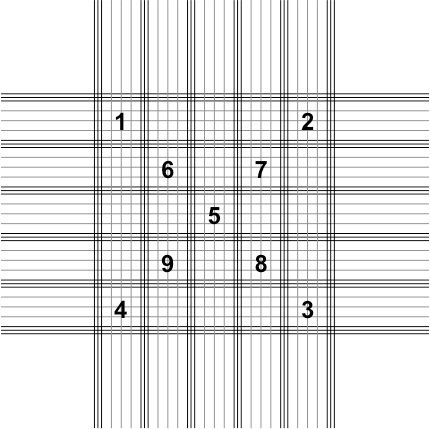
Credit: UF/IFAS Honey Bee Lab
12. Record the total number of spores counted (i.e., the raw spore count) in your notebook if you have chosen to use one.
13. Clean the hemocytometer by wiping with 70% ethanol and a no-lint tissue. Discard the cover slip and the zip-locking bag containing the remaining bee and water mixture.
14. Repeat steps 1–13 for each colony.
Conversion of the Raw Spore Count to the Number of Spores per Bee
The equation below is used to convert each raw spore count into an estimation of number of spores per bee.
(Raw spore count from 5 blocks • 4 million) / # of squares counted = number of spores per bee
Example calculation:
- Let's says that you counted spores from 5 blocks. There are 16 squares within each block (5 • 16 = 80). So the # of squares counted = 80.
- In those 5 blocks you counted a total of 30 spores (the raw spore count 5 blocks)
30 x 4,000,000 = 1,500,000 (or 1.5 million) spores per bee
80
Conclusion
Recent studies have demonstrated that the number of spores per bee can fluctuate depending on bee nutrition and that higher spore counts do not necessarily correlate with honey bee mortality (Zheng et al. 2014; Flemming et al. 2015). However, the Bee Informed Partnership diagnostic services team recommends that beekeepers be concerned when their colonies exceed 1 million spores per bee, even though the number of spores per bee may be somewhat arbitrary. Regardless, accurate sampling methods are a critical component of any integrated pest management program and should be implemented before making timely treatment decisions.
Selected Resources
Cantwell, G. E. 1970. "Standard methods for counting Nosema spores." Am Bee J. 110: 222–223.
Chen, Y, J. D. Evans, L. Zhou, H. Boncristiani, K. Kimura, T-G. Xiao, et al. 2009. "Asymmetrical coexistence of Nosema ceranae and Nosema apis in honeybees." J Invertebr Pathol. 101: 204–209. doi: http://dx.doi.org/10.1016/j.jip.2009.05.012
Fleming JC, Schmehl DR, Ellis JD. 2015. "Characterizing the impact of commercial pollen substitute diets on the level of Nosema spp. in honey bees (Apis mellifera L)." PLoS ONE 10(7): e0132014. doi: http://dx.doi.org/10.1371/journal.pone.0132014
Fries I. 2010. "Nosema ceranae in European honey bees (Apis mellifera)." J Invertebr Pathol. 103: S73–S79. doi: 10.1016/j.jip.2009.06.017
Fries I, Chauzat M-P, Chen Y-P, Doublet V, Genersch E, Gisder S, et al. 2013. Standard methods for Nosema research. In V Dietemann; J D Ellis, P Neumann (Eds) The COLOSS BEEBOOK: Volume II: Standard methods for Apis mellifera pest and pathogen research. J Apic Res 51(5). doi: 10.3896/IBRA.1.52.1.14
Human H. Brodschneider R, Dietemann V, Dively G, Ellis J, Forsgren E, et al. (2013). Miscellaneous standard methods for Apis mellifera research. In V Dietemann; J D Ellis; P Neumann (Eds) The COLOSS BEEBOOK, Volume I: standard methods for Apis mellifera research. J Apic Res 52(4). doi: http://dx.doi.org/10.3896/IBRA.1.52.4.10
Mayack C, Naug D. 2009. "Energetic stress in the honeybee Apis mellifera from Nosema ceranae infection." J Invertebr Pathol. 100: 185–188. doi: 10.1016/j.jip.2008.12.001
Mortensen AN, Schmehl DR, Ellis JD. 2013. Apis mellifera Linnaeus, and subspecies (Insecta: Hymenoptera: Apidae). EENY 568. Gainesville: University of Florida Institute of Food and Agricultural Sciences. http://entnemdept.ufl.edu/creatures/MISC/BEES/euro_honey_bee.htm (March 2019)
Zheng HQ, Lin ZG, Huang SK, Sohr A, Chen YP. 2014. "Spore loads may not be used alone as a direct indicator of the severity of Nosema ceranae infection in honey bees Apis mellifera (Hymenoptera: Apidae)." J Econ Entomol. 107(6): 2037–2044. doi: 10.1603/EC13520