Gastrointestinal nematodes are the most important internal parasites affecting the small ruminant industry in the United States and around the world. They are a major limiting factor of production efficiency in sheep and goats in tropical and subtropical environments. In particular, Haemonchus contortus is the most pathogenic parasite of the abomasum in the Southern and Western United States. According to the USDA National Animal Health Monitoring System, gastrointestinal nematodes were the primary cause of death loss in small ruminants in 2015, resulting in almost 87,000 goat and kid deaths in the US. Similarly, 34,782 lamb and sheep deaths were attributed to gastrointestinal nematodes in 2014.
Traditionally, producers rely on the use of anthelmintic drugs to control internal parasites. However, widespread anthelmintic resistance has encouraged the development of alternative strategies. Development and use of population-specific genomic tools in selection to improve resistance to gastrointestinal nematodes represents an alternative approach. Breeding parasite-resistant sheep that are less dependent on the use of anthelmintics improves sustainability and profitability of sheep and goat farming systems in hot and humid regions of the United States. The main benefits of using selection to enhance parasite resistance in sheep and goat systems are that animals can be selected at an early age, the genetic change is transmitted to the following generations, and infection is decreased, leading to additional benefits for susceptible animals grazing in the same flock. Besides phenotypic markers such as fecal egg count, genetic markers can be included in sheep and goat breeding programs to identify individuals with enhanced resistance at an early age. Genetic markers are small pieces of DNA, or single nucleotide polymorphisms (SNPs), with a known location on the animal’s genome and association with a particular gene or trait. Thus, if SNPs that control resistance to gastrointestinal nematodes are detected, they can be used in selection to identify genetically superior rams and bucks early in life. Moreover, selection for enhanced resistance creates unique genetic patterns in the genome of these species that can be traced back.
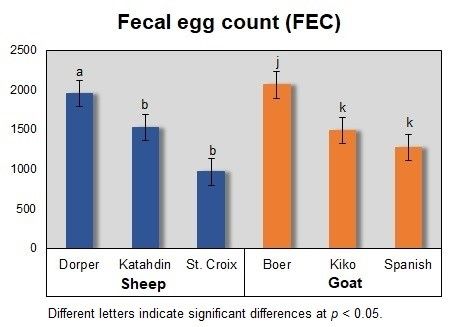
A research project funded by the USDA National Institute of Food and Agriculture 1890 Institution Capacity Building Grant Program was designed to explore the genetics of resistance to gastrointestinal parasites in sheep and goats. For this project, phenotypic records from commercial farms in Arkansas, Kansas, and Missouri, and the American Institute for Goat Research at Langston University Oklahoma were used. The study included 144 male sheep and 144 male goats. Dorper (n = 47), Katahdin (n = 57), and St. Croix (n = 40) sheep and Boer (n = 51), Kiko (n = 46), and Spanish (n = 47) goat breeds were used to construct the phenotype-genotype databases. Figure 1 presents a bar graph of fecal egg count to illustrate the phenotypic variation across species and among breeds in the study.
Credit: UF/IFAS
Several traits were measured in sheep and goats, including fecal egg count, hematocrit, average daily gain, and immunoglobulin levels. Genomic DNA obtained from blood samples of these species was utilized for sequencing using a targeted approach. Genotypes from SNP data were then utilized to perform the identification of potential genetic markers associated with resistance to gastrointestinal nematodes and evaluate the genetic patterns for resistance.
Sheep populations presented lower fecal egg count and stronger humoral response than goat breeds. For sheep, St. Croix had the lowest fecal egg count among breeds. Among goats, the Spanish breed had the lowest fecal egg count. Results from genotype analysis revealed that for sheep, SNPs located on chromosomes 1 and 2 were associated with average daily gain (ADG), immunoglobulin M (IgM), and fecal egg count. For goats, SNPs located on chromosomes 3 and 22 were associated with ADG and IgM. Although different SNPs were associated with resistance, significant markers within IL12RB2 gene were identified in both species.
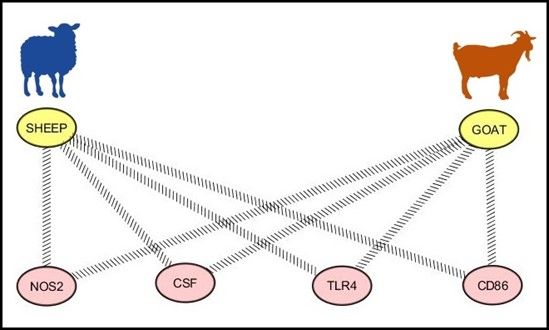
Moreover, shared immune response mechanisms between sheep and goats were identified in the genome of these species (Figure 2). The NOS2, CSF, TLR4, and CD86 genes were observed under selection in both species. These genes are key modulators of immune responses, and active players of parasite-antigen recognition by immune response cells such as dendritic cells and T-cells. The conserved mechanisms of protection against H. contortus are most likely to be potential targets in the development of alternative nematode control strategies in both species, because these mechanisms can be widely applied in production systems. For this reason, future efforts will focus on validation of the observed results and the unraveling of the genetic mechanisms used for controlling H. contortus or other gastrointestinal parasites in sheep and goats.
Credit: UF/IFAS